|
Cross-Referenced Tables are just tab-delimited flat files, which encapsulate data in an object hierarchy with arbitrary attributes and relationships. |
1. General Description
XRT organizes data into different classes (e.g. gene, transcript, clone, mutation, disease etc.); each class has many attributes to describe the properties of its elements. The XRT file Gene.xrt below contains four elements, each with six attributes; the first line specifies the attribute names, while the following lines contain the actual attribute values for elements.
Three attributes (ID, P_ID, C_ID) have special meaning to the system. The ID field (mandatory to all elements) stores the unique identifier of each data element; P_ID / C_ID are used for more complex parent / child relationships between data elements.
Example1: XRT file Gene.xrt keeps the elements of Gene class
|
Example2: XRT file Transcipt.xrt keeps the elements of Transcript class
|
Normally, one class is represented by one XRT file, containing all of its attributes; however, this is not mandatory while data being drawn from multiple sources, in this case, one class can have multiple XRT files, each keeps a subset of attributes of that class.
Class name is specified by the name of XRT file, the string before the first dot (.) of the file name will be the class name. For example, both XRT files Gene.xrt and Gene.OMIM.xrt are for the Gene class.
Gene.OMIM.xrt looks like this:
|
The first line of XRT file has to be the attributes of its class, attribute names are delimited by a tab, and the first attribute has to be ID. The following lines contain the actual attribute values for elements of the class, one element each line. Every element (or data entry) must have a global unique identifier (kept in the ID attribute, again, ID attribute is mandatory to any XRT table), for example, GA0001 is the unique identifier for Gene ABP1, and T02767 is the unique identifier for a transcript.
Attribute can have single value, no value (empty), or multiple values. In the case of multiple values, use '&;' (initially, '\t' was used) to separate them.
Elements in different classes may be related to each other, for example, one gene has one or more transcripts. In the XRT files above, you can see gene GA0001 has two transcripts (T00001 and T02767), gene GA0003 has one (T00005).
Relationships are kept by P_ID and C_ID (stands for parent element ID and child element ID respectively). When you specify the related element of the current element, you always have to indicate the class of the related element as well, such as: P_ID/Gene and C_ID/LoucsLink, meaning the parent class is Gene and the child class is LocusLink respectively.
Relationship can be one to one, one to many or many to many, i.e. one element can have one or more related elements, and/or multiple elements can share one related element. This flexibility makes XRT very powerful to model complex biological data.
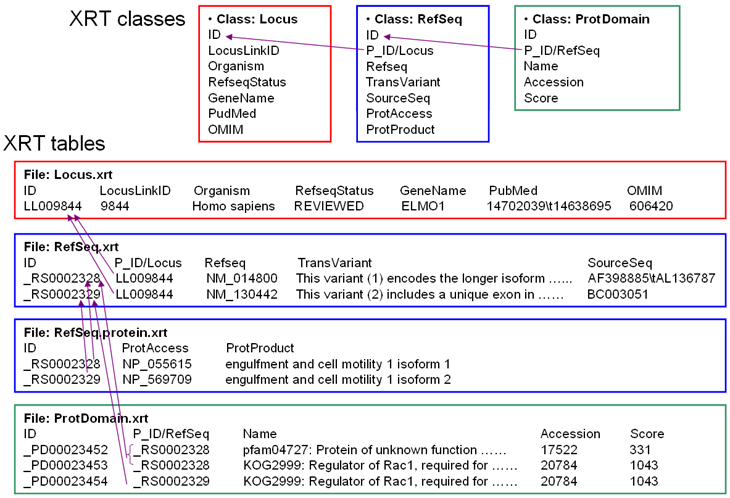
Below is an example showing three related classes, the relationship can be summarized as: one Locus has one or more Refseq, one Refseq has one or more ProtDomain. Please note that the Refseq class is represented by two XRT files (Refseq.xrt and Refseq.protein.xrt).